GUI Usage
SarcAsM’s GUI application uses Napari image browser as image viewer. Check out this page for a primer on Napari’s GUI.
Starting the App
There are two main ways to start the SarcAsM GUI:
1. Standalone Applications (Recommended for ease of use):
Pre-built applications for Windows (.exe) and macOS (.app) are available for download directly from the GitHub Releases page.
This method does not require a separate Python installation.
Note:
These standalone applications are early versions and may take a significant amount of time to start up initially.
They are built using Python 3.11.
The Windows version currently only utilizes the CPU and does not support CUDA GPU acceleration. For high-performance needs, consider using the Python API.
2. From your Python Environment (Recommended for developers or API users):
If you have installed SarcAsM into a Python environment (e.g., via pip or conda):
First, remember to activate your installation environment:
conda activate sarcasm-env # Or your specific environment name
Then, to start the app, run this command:
python -m sarcasm_app
Using the App
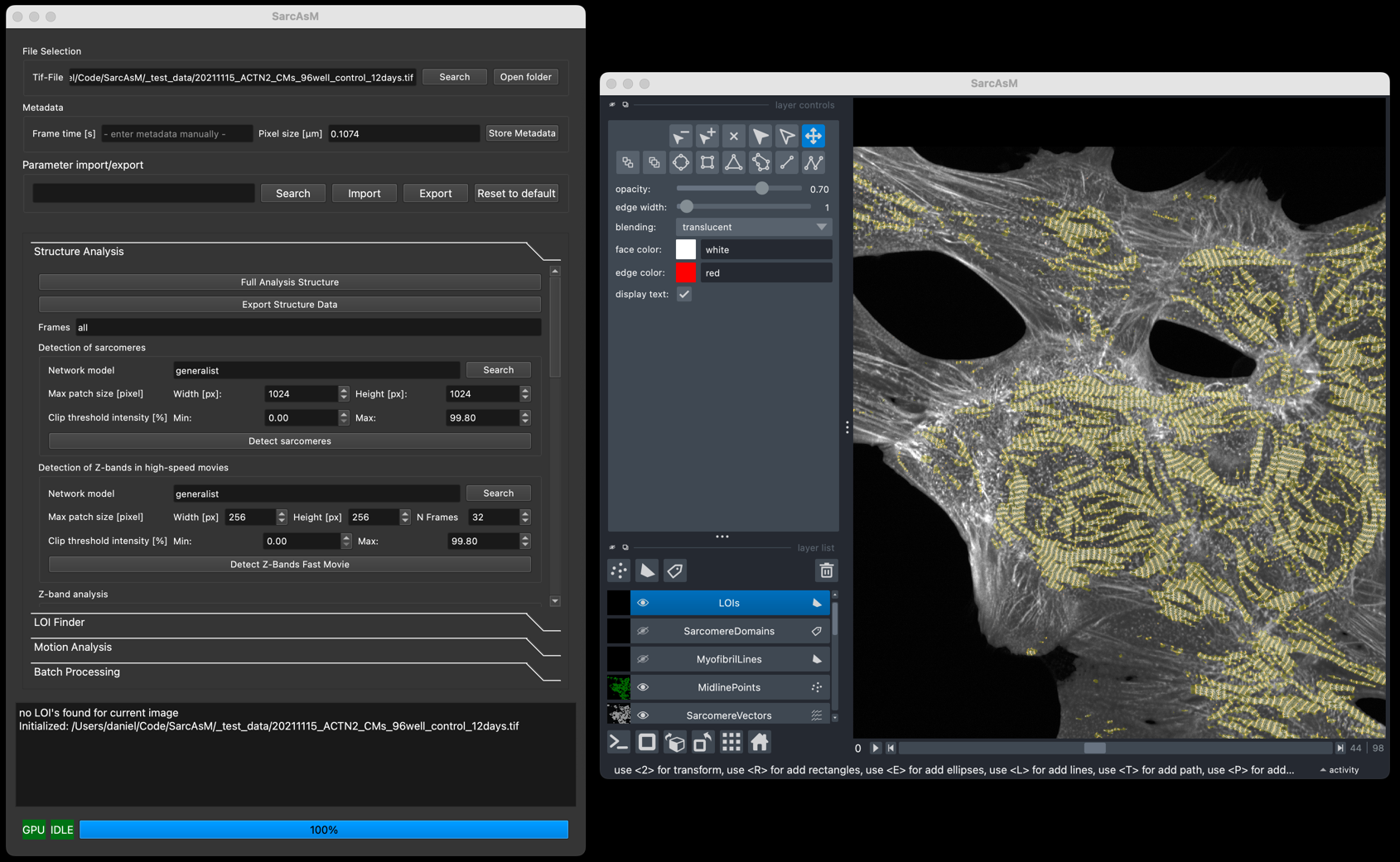
The SarcAsM GUI integrates controls (left panel) with a Napari image viewer (right panel). Follow the steps in the control panel sections for analysis.
Initial Steps:
Load File: Use Search in File Selection to load a grayscale TIFF file.
Set Metadata: In Metadata, check the Pixel size [µm]. If the field is red or incorrect, enter the correct value and press Store Metadata.
Analysis Sections:
Proceed through the collapsible sections in the control panel:
Parameter import/export: Load/save analysis settings or reset to defaults.
Structure Analysis: Analyze sarcomere structure.
LOI Finder: Find and/or draw Lines of Interest (requires structure results).
Motion Analysis: Analyze sarcomere motion in LOIs.
Batch Processing: Process multiple files automatically.
Using Parameters:
Tooltips: Hover over parameter names/fields for descriptions.
If needed, optimize settings within each section before running the corresponding analysis step (e.g., clicking Detect sarcomeres).